Higher-order Singular Value Decomposition
In multilinear algebra, there does not exist a general decomposition method for multi-way arrays (also known as N-arrays, higher-order arrays, or data-tensors) with all the properties of a matrix singular value decomposition (SVD). A matrix SVD simultaneously computes
(a) a rank-R decomposition and
(b) the orthonormal row/column matrices.
These two properties can be captured separately by two different decompositions for multi-way arrays.
Property (a) is extended to higher order by a class of closely related constructions known collectively as CP decomposition (named after the two most popular and general variants, CANDECOMP and PARAFAC). Such decompositions represent a tensor as the sum of the n-fold outer products of rank-1 tensors, where n is the dimension of the tensor indices.
Property (b) is extended to higher order by a class of methods known variably as Tucker3, N-mode SVD, and N-mode principal component analysis (PCA). (This article will use the general term “Tucker decomposition”.) These methods compute the orthonormal spaces associated with the different axes (or modes) of a tensor. The Tucker decomposition is also used in multilinear subspace learning as multilinear principal component analysis. This terminology was coined by P. Kroonenberg in the 1980s, but it was later called multilinear SVD and HOSVD (higher-order SVD) by L. De Lathauwer.
Historically, much of the interest in higher-order SVDs was driven by the need to analyze empirical data, especially in psychometrics and chemometrics. As such, many of the methods have been independently invented several times, often with subtle variations, leading to a confusing literature. Abstract and general mathematical theorems are rare (though see Kruskal with regard to the CP decomposition); instead, the methods are often designed for analyzing specific data types. The 2008 review article by Kolda and Bader^[2]^ provides a compact summary of the history of these decompositions, and many references for further reading.
The concept of HOSVD was carried over to functions by Baranyi and Yam via the TP model transformation. This extension led to the definition of the HOSVD based canonical form of tensor product functions and Linear Parameter Varying system models and to convex hull manipulation based control optimization theory, see TP model transformation in control theories.
CP decomposition
Main article: CP decomposition
Definition
A CP decomposition of an N-way array X, with elements  ,
is
,
is

where
 denotes the tensor product. The
R tensors
denotes the tensor product. The
R tensors
 (known as simple tensors, rank-1 tensors, dyads, or, in quantum
mechanics, product
states) are constructed from the rN vectors
(known as simple tensors, rank-1 tensors, dyads, or, in quantum
mechanics, product
states) are constructed from the rN vectors  .
With indices, this is
.
With indices, this is

where
 is the i-th element of the vector
is the i-th element of the vector
 ,
etc.
,
etc.
Tucker decomposition
Main article: Tucker decomposition
History
In 1966, L. Tucker proposed a decomposition method for three-way arrays (referred to as a 3-mode “tensors”) as a multidimensional extension of factor analysis. This decomposition was further developed in the 1980s by P. Kroonenberg, who coined the terms Tucker3, Tucker3ALS (an alternating least squares dimensionality reduction algorithm), 3-Mode SVD, and 3-Mode PCA. In the intervening years, several authors developed the decomposition for N-way arrays. Most recently, this work was treated in an elegant fashion and introduced to the SIAM community by L. De Lathauwer et al. who referred to the decomposition as an N-way SVD, multilinear SVD and HOSVD.
Definitions
Let the SVD of a real matrix be  ,
then it can be written in an elementwise form as
,
then it can be written in an elementwise form as

 and
and
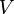 give, in a certain sense optimal, orthonormal basis for the column and
row space,
give, in a certain sense optimal, orthonormal basis for the column and
row space,
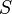 is diagonal with decreasing elements. The higher-order singular value
decomposition (HOSVD) can be defined by the multidimensional
generalization of this concept:
is diagonal with decreasing elements. The higher-order singular value
decomposition (HOSVD) can be defined by the multidimensional
generalization of this concept:

where the ![U\^{(n)} = [u\^{(n)}_{i,j}]_{I_n \times
I_n}](http://upload.wikimedia.org/math/6/d/5/6d53b3015286aaae8b1fcf30efebd9e7.png) matrices and the
matrices and the ![\mathcal{S} = [s_{j_1,\dots,j_N}]_{I_1 \times
I_2 \times \cdots \times
I_N}](http://upload.wikimedia.org/math/d/1/a/d1a449f44327b3a4e898d443299c3174.png) core tensor should satisfy certain requirements (similar ones to the
matrix SVD), namely
core tensor should satisfy certain requirements (similar ones to the
matrix SVD), namely
- Each
 is an orthogonal
matrix.
is an orthogonal
matrix. - Two subtensors of the core tensor
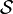 are orthogonal i.e.,
are orthogonal i.e.,  if
if 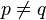 .
. - The subtensors in the core tensor
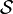 are ordered according to their Frobenius
norm, i.e.
are ordered according to their Frobenius
norm, i.e.
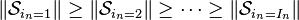 for n = 1, …, N.
for n = 1, …, N.
Notation:

Algorithm
The HOSVD can be built from several SVDs, as follows:
- Given a tensor
 ,
construct the mode-k flattening
,
construct the mode-k flattening
 .
That is, the
.
That is, the  matrix that corresponds to
matrix that corresponds to
 .
. - Compute the singular value
decomposition
 ,
and store the left singular vectors
,
and store the left singular vectors
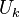 .
. - The core tensor
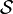 is then the projection of
is then the projection of
 onto the tensor basis formed by the factor matrices
onto the tensor basis formed by the factor matrices
 ,
i.e.,
,
i.e., 
Applications
Main applications are extracting relevant information from multi-way arrays. Used in factor analysis, face recognition (TensorFaces), human motion analysis and synthesis.
The HOSVD has been successfully applied to signal processing and big data, e.g., in genomic signal processing.and a tensor GSVD.
A combination of HOSVD and SVD also has been applied for real time event detection from complex data streams (multivariate data with space and time dimensions) in Disease surveillance.
It is also used in tensor product model transformation-based controller design. In multilinear subspace learning, it is modified to multilinear principal component analysis for gait recognition.
In machine learning, the CP-decomposition is the central ingredient in
learning probabilistic latent variables models via the technique of
moment-matching. For example, let us consider the multi-view
model which is a
probabilistic latent variable model. In this model, we posit the
generation of samples as follows: there exists a hidden random variable
that is not observed directly, given which, there are several
conditionally
independent
random variables known as the different “views” of the hidden variable.
For simplicity, let us say we have three symmetrical views
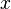 of a
of a
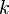 -state
categorical hidden variable
-state
categorical hidden variable
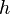 .
Then the empirical third moment of this latent variable model can be
written as:
.
Then the empirical third moment of this latent variable model can be
written as: ![T=\sum_{i=1}\^{k}Pr(h=k) E[x|h=k]\^{\otimes
3}](http://upload.wikimedia.org/math/1/b/0/1b02e1da9ddc7a8d5a2ef403f8e6ad83.png) .
.
In applications such as topic
modeling, this can be
interpreted as the co-occurrence of words in a document. Then the
eigenvalues of this empirical moment tensor can be interpreted as the
probability of choosing a specific topic and each column of the factor
matrix
![E[x|h=k]](http://upload.wikimedia.org/math/9/c/9/9c9e95c7547d3335723ade46cef2babc.png) corresponds to probabilities of words in the vocabulary in the
corresponding topic.
corresponds to probabilities of words in the vocabulary in the
corresponding topic.
References
- Kruskal, J. B. (1989). “Rank, decomposition, and uniqueness for 3-way and N-way arrays”. In R. Coppi & S. Bolasco (Eds.), Multiway data analysis (pp. 7–18). Amsterdam: Elsevier. [ PDF ].
- Kolda, Tamara G.; Bader, Brett W. “Tensor Decompositions and Applications”. SIAM Rev. 51: 455–500 (46 pages). doi:10.1137/07070111X. CiteSeerX: 10.1.1.153.2059.
- P Baranyi (April 2004). “TP model transformation as a way to LMI based controller design”. IEEE Transaction on Industrial Electronics 51 (2): 387–400. doi:10.1109/tie.2003.822037.
- P Baranyi and D. Tikk and Y. Yam and R. J. Patton (2003). “From Differential Equations to PDC Controller Design via Numerical Transformation”. Computers in Industry, Elsevier Science 51: 281–297. doi:10.1016/s0166-3615(03)00058-7.
- P Baranyi and L. Szeidl and P. Várlaki and Y. Yam (July 3–5, 2006). Definition of the HOSVD-based canonical form of polytopic dynamic models. Budapest, Hungary. pp. 660–665.
- Ledyard R. Tucker](https://en.wikipedia.org/wiki/Ledyard_R._Tucker “Ledyard R. Tucker”) (September 1966). “Some mathematical notes on three-mode factor analysis”. Psychometrika 31 (3): 279–311. doi:10.1007/BF02289464.
- P M. Kroonenberg (1983). “Three-mode principal component analysis: Theory and applications”. DSWO Press, Leiden.
- Lieven De Lathauwer, Bart De Moor and Joos Vandewalle (April 2000). “A multilinear Singular Value Decomposition”. SIAM Journal on Matrix Analysis 21 (4): 1253–1278.
- L Omberg, G. H. Golub and O. Alter (November 2007). “A Tensor Higher-Order Singular Value Decomposition for Integrative Analysis of DNA Microarray Data From Different Studies”. PNAS 104 (47): 18371–18376. doi:10.1073/pnas.0709146104. PMC 2147680. PMID 18003902.
- L Omberg, J. R. Meyerson, K. Kobayashi, L. S. Drury, J. F. X. Diffley and O. Alter (October 2009). “Global Effects of DNA Replication and DNA Replication Origin Activity on Eukaryotic Gene Expression”. Molecular Systems Biology 5: 312. doi:10.1038/msb.2009.70. PMC 2779084. PMID 19888207. Highlight.
- C Muralidhara, A. M. Gross, R. R. Gutell and O. Alter (April 2011). “Tensor Decomposition Reveals Concurrent Evolutionary Convergences and Divergences and Correlations with Structural Motifs in Ribosomal RNA”. PLoS ONE 6 (4): e18768. doi:10.1371/journal.pone.0018768. Highlight.
- S P. Ponnapalli, M. A. Saunders, C. F. Van Loan and O. Alter (December 2011). “A Higher-Order Generalized Singular Value Decomposition for Comparison of Global mRNA Expression from Multiple Organisms”. PLoS One 6 (12): e28072. doi:10.1371/journal.pone.0028072. Highlight.
- P Sankaranarayanan, T. E. Schomay, K. A. Aiello and O. Alter (April 2015). “Tensor GSVD of Patient- and Platform-Matched Tumor and Normal DNA Copy-Number Profiles Uncovers Chromosome Arm-Wide Patterns of Tumor-Exclusive Platform-Consistent Alterations Encoding for Cell Transformation and Predicting Ovarian Cancer Survival”. PLoS One 10 (4): e0121396. doi:10.1371/journal.pone.0121396. AAAS EurekAlert! Press Release and NAE Podcast Feature.
- Hadi Fanaee-T and João Gama (May 2015). “EigenEvent: An algorithm for event detection from complex data streams in Syndromic surveillance”. Intelligent Data Analysis 19 (3).
- Haiping Lu, K.N. Plataniotis and A.N. Venetsanopoulos, “A Survey of Multilinear Subspace Learning for Tensor Data”, Pattern Recognition, Vol. 44, No. 7, pp. 1540–1551, Jul. 2011.
- H Lu, K. N. Plataniotis, and A. N. Venetsanopoulos, “MPCA: Multilinear principal component analysis of tensor objects,” IEEE Trans. Neural Netw., vol. 19, no. 1, pp. 18–39, Jan. 2008.
- Anandkumar, Animashree; Ge, Rong; Hsu, Daniel; Kakade, Sham M; Telgarsky, Matus (2014). “Tensor decompositions for learning latent variable models”. The Journal of Machine Learning Research 15 (1): 2773–2832.